Alzheimer's and Dementia Research
In This Section
- Scaling Evidenced-Based Care: Early Wins as New Treatments Emerge
- Big Data Power: From Clinics to Big Data to Test Tubes and Back to the Clinics
- Discovering Promising Treatments: Closing the Loop From Clinic to Research to Clinic
- Innovation - New Blood Tests: Exosomes
- Innovation - Brain Scans: Diagnosing and Predicting
- Innovation: Stem Cell Models
Scaling Evidenced-Based Care: Early Wins as New Treatments Emerge
The Richman Center analyzes medical records of large numbers of patients receiving primary care from Johns Hopkins Community Physicians (JHCP), as well as medical records of Memory and Alzheimer’s Treatment Center (MATC) patients when they first present for treatment. One goal is to identify subgroups predicted to develop ADRD in the future or with worse outcomes based on their course after diagnosis (prognosis). Another goal is to define subgroups most likely to respond to the medications available and approved by the Food and Drug Administration (e.g., Aricept, Exelon, Nameda, Leqembi) for Alzheimer’s, as well as a series of other medication and nonmedication interventions proved to be helpful.
A priority for the Richman Center is to achieve early wins that help patients in the next few years while more intensive cure-oriented studies unfold over longer time periods. Using “big data” analyses, the center will develop algorithms that predict which JHCP and MATC patients do worse after diagnosis in order to provide them with more intensive home-based interventions targeting their life quality and providing support to their caregivers. The delivery of these achievements will be anchored by the Johns Hopkins Maximizing Independence at Home (MIND at Home) system of care for people with cognitive disorders, which combines pharmacologic, behavioral and a variety of other interventions to improve patient and caregiver outcomes, reduce health care use and improve quality of care.
Big Data Power: From Clinics to Big Data to Test Tubes and Back to the Clinics
The Richman Center will establish and follow large cohorts of well-characterized MATC and primary care patients in partnership with JHCP, which operates over 30 primary care clinics in the Baltimore-Washington region.
For patients with brain imaging studies conducted at MATC, digital results will be linked to medical records and analyzed using “big data” methods. The center will obtain blood from patients for EV and stem cell studies in a unique translational project aimed at characterizing biologic subtypes of the clinical phenotype of ADRD.
Because these biomarkers will be developed from clinically well characterized patients, it will be possible to link clinical and digital brain imaging information with blood test findings in a highly novel fashion to develop and test hypotheses about how best to subtype AD in ways that will be therapeutically relevant.
Knowledge derived from these studies will inform future studies of the human cohorts (e.g., by development of new biomarkers of specific AD subtypes), thus closing the loop from clinic to test tube and back to the clinic.
Discovering Promising Treatments: Closing the Loop From Clinic to Research to Clinic
There is currently no highly effective treatment for Alzheimer’s disease. Many medications have failed in trials. Identification of new treatments is urgently needed. Most efforts aiming at disease modification are focused on the amyloid hypothesis that AD is caused by abnormal buildup of the protein amyloid around brain cells, resulting in cell death. FDA is in the process of approving a new class of meds that effectively remove amyloid from the brain but have only minimal benefit for patients while carrying notable risks of brain swelling and bleeds. Recent studies showed that multiple disease processes are involved in the onset and progression of ADRD, including processes that affect brain blood vessels, which for some — but not all — people may play a big role.
Richman Center investigators are applying a novel MRI-based brain imaging technique that enables measurement of brain vessel reactivity (CVR) in response to an inhaled mixture of room air with carbon dioxide. Healthy brain vessels respond by widening in order to allow more oxygen flow, while a brain with impaired blood vessels, such as the brains of some patients with MCI or AD, do not react accordingly, resulting in chronically decreased oxygen flow when under stress.
Whole brain CVR in participants with normal and impaired cognition — the MRI image shows that oxygen flow (red areas) is less in people with mild cognitive impairment (MCI) and/or AD due to decreased CVR
The Richman Center is funding its first of what are projected to be many treatment discovery projects to determine if using this novel MRI-based CVR technique can help identify a subgroup of people with MCI for whom vascular risk factors play a major role. The goal then is to try to improve CVR in that subgroup by repurposing a widely available safe medicine.
Innovation - New Blood Tests: Extracellular Vesicles (EVs)
Despite the devastating impact of Alzheimer’s disease on patients, their families and society, there is still no cure. Interest has grown in developing treatments that target multiple processes leading to AD, and doing so at earlier stages of brain disease when brain damage may still be reversible. Although the early stages of AD are promising targets for new treatments, they are more difficult to diagnose clinically than later stages. This is especially true for pre-clinical stages of the disease during which, by definition, there are no clinical symptoms and destructive disease processes operate silently.
A variety of biological markers (biomarkers) have been proposed for earlier detection, including during pre-clinical stages of AD. Biofluids such as cerebrospinal fluid (CSF — the fluid that surrounds and cushions the brain) and blood contain molecules that may allow us to determine who is at greater risk for developing AD in the future, or to monitor disease progression in those who already have AD. Some biomarkers are proteins, such as amyloid-beta and tau, or protein fragments that are signature abnormalities of the disease. Both are abnormally modified and form aggregates (“clumps”) in the brain of patients with AD. Damaged neurons and other brain cells also release molecules and cell particles that can be detected outside the brain.
In recent years, extracellular vesicles (EVs), a term referring to very, very, very small particles that are surrounded by a membrane, have attracted considerable attention. These tiny bubblelike structures are released from all cells in the body and are usually only a millionth the size of the parent cell. Their small size and other properties allow EVs to cross boundaries (such as the blood-brain barrier) and move around in the blood circulation.
Importantly, they form a kind of postal system in the blood circulation, with a “return address” of their cell of origin, like a neuron or other brain cell type. By picking EVs from blood with a brain cell return address, we can select only the “mail” we want to read, giving us a much more specific way to eavesdrop on the communications and developing abnormalities in the brain. Richman Center scientists are trying to learn more about this communication system and how it can be used to diagnose patients with as early stage disease as possible, and to identify those with particular abnormalities that may not be shared by all patients with AD. This is a major precision medicine goal for AD. These signals may also give us clues about how to intervene for individual patients to stop or reverse disease abnormalities in a way tailored to how the disease operates. Finally, we can use these signals to know if an experimental treatment works as it is supposed to do at the level of brain cells and molecules.
Innovation - Brain Scans: Diagnosing and Predicting
There have been substantial advances in the use of brain imaging to diagnose, predict and track AD and MCI. However, research advances in imaging have not moved into the routine clinical diagnosis of AD, during which brain scans most often serve as a “rule out” for major structural lesions as the potential cause of cognitive losses. Translating these significant advances in digital neuroimaging research into routine clinical care is needed. The MATC at Johns Hopkins, and other Johns Hopkins Medicine clinical facilities, are invaluable resources to study clinically based neuroimaging markers of cognitive disorders. At the MATC, patients receive a comprehensive cognitive, neuropsychiatric and functional evaluation, which often includes MRI of the brain. Patients are diversely diagnosed (ranging from no cognitive impairment to severe dementia), and a majority of them are followed over time.
Richman Center scientists are constructing a database linking detailed clinical information from patients evaluated in the MATC with automated quantitative analysis of their 3T brain MR scans (figure 1). Image processing will include analyses of regional gray matter volumes and white matter structural integrity measures using DTI, including cortical and limbic structures. This database can be used widely at Johns Hopkins and anywhere MRIs scans are taken for automated analysis that will improve diagnosis, prognosis and targeting of medication treatments.
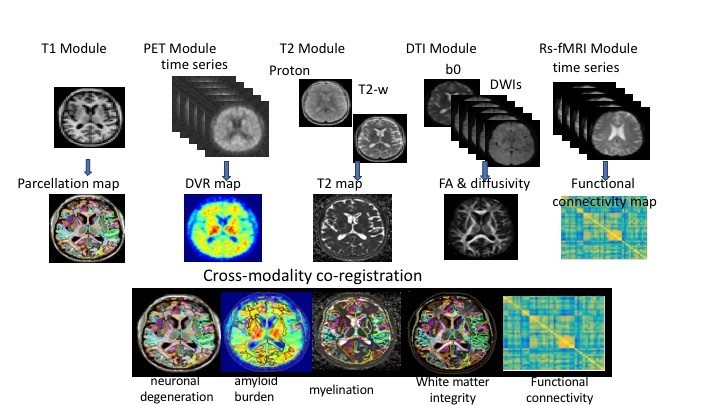 Figure 1. Automated Imaging Pipeline
Figure 1. Automated Imaging PipelineThe project’s eventual goal is to establish the clinical utility of automated analytic procedures for brain MRIs in the diagnosis, prognosis and treatment of patients with cognitive disorders. Associations between brain structure and function and cognitive, behavioral, medical and demographic variables can be made to gain insights into the complex interactions of a heterogeneous group of people over time. (Fig. 2)
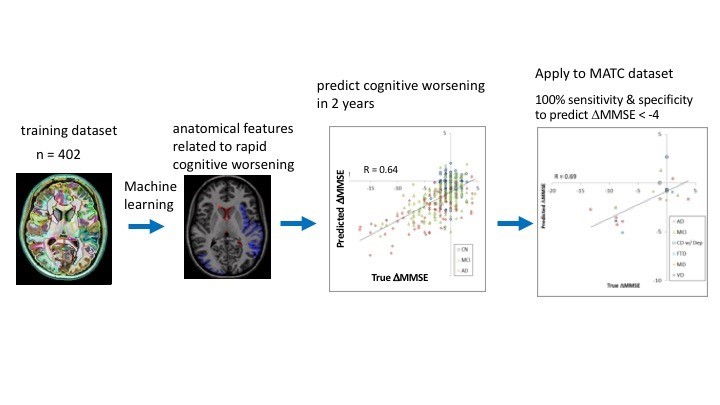 Figure 2. Cloud-based brain MRI segmentation and parcellation system using a machine learning algorithm predicts the change in cognitive scores in two years.
Figure 2. Cloud-based brain MRI segmentation and parcellation system using a machine learning algorithm predicts the change in cognitive scores in two years.Innovation: Stem Cell Models
Alzheimer’s disease is one of the most devastating brain diseases characterized by progressive cognitive impairments, including memory loss and behavioral change. Hallmark neuropathologies include the extracellular deposition of β-amyloid (Aβ) in senile plaques and the intracellular aggregation of tau protein in the form of neurofibrillary tangles.
The problem of how to study the AD brain disease in the lab for the purposes of treatment development was initially tackled by “transgenic” mouse models. While the models advanced the study of AD, they failed to lead to effective treatments because of species-specific differences in brain development. Human models of the AD brain disease are needed to advance treatment development.
The discovery of human induced pluripotent stem cells (iPSCs) derived from a person’s blood now provides unprecedented opportunities for AD treatment development. iPSCs from individuals can be differentiated in the lab into all brain functional cell types. iPSCs also capture unique genetic information, thus serving as excellent human, personalized models to understand molecular pathogenesis and develop treatments targets.
Supported by the Richman Family Precision Medicine Center of Excellence, the Machairaki Laboratory focuses on the application of hiPSCs as models to explore molecular mechanisms underlying AD and other brain diseases, leading to the development of novel therapeutics. In the past, the lab generated iPSCs from human skin cells of people carrying the presenilin (PS1) DNA mutations that cause familial AD. Using cutting edge reprogramming strategies, these skin cells were differentiated into functional neurons that expressed the pathogenic gene and produced amyloid. The lab has now extended its techniques and can reproducibly derive iPSCs from blood cells of individuals, which are now routinely being differentiated into neurons and other brain cells.
Even though we can efficiently and reproducibly generate, from blood, specific brain cell types using two-dimensional (2D) cultures, these methods are unlikely to recapitulate the neural tissue organization in vivo. This problem has been tackled with the advent of cerebral organoids — i.e., 3D culture systems derived from human iPSCs that recapitulate distinct architectures of the human brain, composed of progenitor and neural cells, organized in layers almost as in the native brain. The use of cerebral organoids closely mimics the cellular environment of living organisms.
A decades-long aspiration in the field of AD is now within reach: the ability to model the AD brain disease of individuals using iPSCs-derived 2D and 3D human models. Using these systems, cells can grow safely in two or three dimensions and, more importantly, can more closely mimic the cellular environment of living organisms under pathological or normal conditions.
As part of the Richman Center, the Machairaki Lab focuses on a unique personalized medicine approach involving the development of hiPSC models for the characterization of AD biologic subtypes. To reach this goal, we obtain blood from patients with or at risk for AD, as well as from comparison groups followed by generation of each individual’s iPSC lines. These cell lines are differentiated to each person’s brain cells, namely different types of neurons, astrocytes and microglial cells, as well as organoids that are being used as platforms for biomarker discovery and drug screening (see the chart).
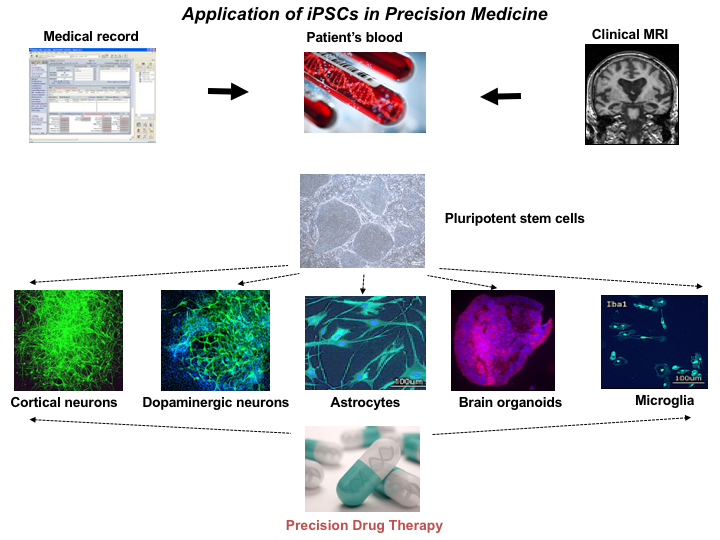 Figure 1. Application of iPSCs in Precision Medicine
Figure 1. Application of iPSCs in Precision MedicineOur in vitro iPSC models have an unprecedented level of cellular resolution and great experimental versatility. These features offer unique opportunities for testing drugs at the single person level, developing disease biomarkers, and asking questions about cellular and molecular aspects of pathogenesis. Human iPSCs-derived brain cells can provide a unique platform to model AD on a patient-by-patient basis. Differentiation of iPSCs from large numbers of patients with AD will allow classification of the patients into subtypes depending on their response to drug tests.
The advent of iPSC-derived individual person models of the AD brain disease will also significantly improve the way clinical trials are performed in the future. Testing a drug investigated in a clinical trial will be first studied in a patient’s iPSC-derived brain cells, leading to fast selection of a subgroup of patients who are likely to respond positively, accelerating the drug under study to the next phases of the trial (Figure 2).
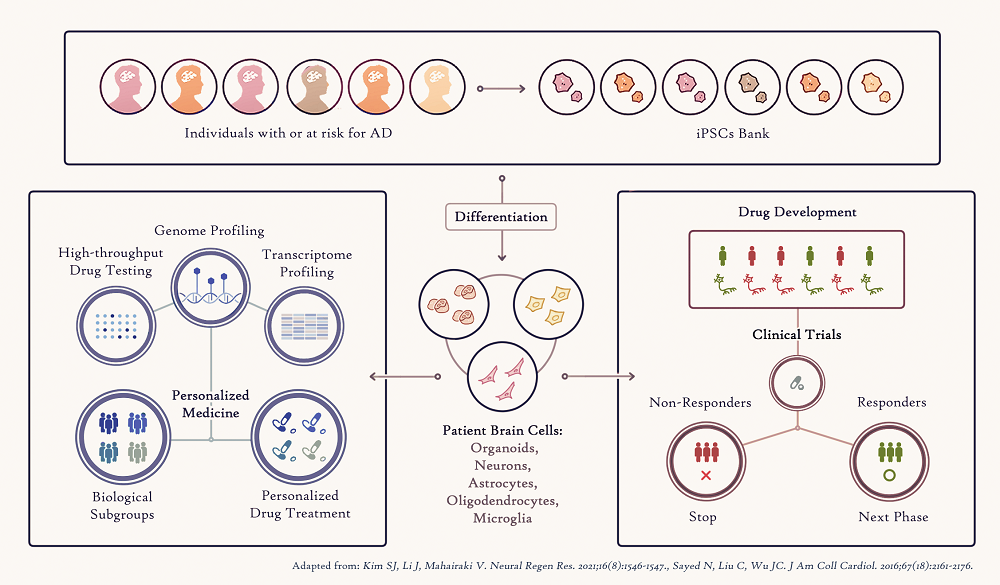 Figure 2. Applications of human iPSCs to Precision Medicine
Figure 2. Applications of human iPSCs to Precision MedicineFinally, iPSCs provide the opportunity to study the exact genomic (DNA) information of each patient, enabling study of the genetic basis of AD and how it can influence response to a drug treatment. Combining the genomic analysis of numerous Alzheimer’s disease iPSC-brain cells in a dish and drug screening assays in these cells will help identify patient-specific genetic variations responsible for effective person-specific drug treatment that could then serve as biomarkers. The Richman Center is also supporting the Avramopoulos Genetics Lab to explore ways to expand our information input from rare but highly deleterious Alzheimer’s-causing DNA mutations to less deleterious but very common disease-causing variation through combining them as biology informed polygenic risk scores in prediction models.
