Past Studies
Home | Contact Us | Our People | Active Studies | Tools | Researchers | Participants
Early Identification of Preclinical Coronary Atherosclerosis in High Risk Families
GeneSTAR has over 25 years of experience in assessing noninvasive measures of occult preclinical coronary disease including perfusion imaging and coronary angiography. We have shown that the results of perfusion imaging are potent predictors of clinical coronary disease, and of its nature and severity. We have extended this work to CT angiography, examining both calcified and noncalcified plaque, extending the work beyond phenotype-phenotype associations to genomics. More recently we have begun to examine areas of inflammation in the coronary arteries using positron emission tomography with a new agent.
CT Angiography Studies
The prime goal is to compare an innovative coronary artery imaging technology with a standard imaging method in the detection of very early coronary disease in people who are at high risk but who have not had symptoms or a clinical coronary disease event.
Coronary atherogenesis begins well before manifest clinical coronary disease (CAD). If occult CAD and its extent could be accurately identified during the preclinical phase, targeted intensive preventive therapies could be tested that may mitigate CAD events. In this study we apply a comparative effectiveness research paradigm to a susceptible population of asymptomatic first degree adult relatives of persons with known premature CAD, all easily identifiable by family history, with a CAD risk that is 2-5 times that of the general population
The objective is to advance personalized preventive medicine based on early detection of coronary disease and its forms. We compare CT CAC and MDCTA for identifying latent preclinical CAD in GeneSTAR subjects who represent appropriate targets for intensive preventive therapies. Our aims are to:
- evaluate both the extent of calcified and noncalcified coronary plaque using MDCTA, and coronary calcium score (CAC) in 1000 22 to 75 year old apparently healthy siblings and adult offspring of probands with known premature CAD (< 60 years of age) from the GeneSTAR study.
- determine the relationship of traditional risk factors (Framingham Global Risk Score), and hsCRP to outcomes to determine the optimal identification of persons with significant preclinical CAD, and
- determine whether the addition of known genetic risk variants for CAD improve selection of subjects for CT or MDCTA.
Data is being used to generate reference values for plaque volumes and to determine the value of MDCTA as a method to track responsiveness to therapy in primary prevention trials.
Done in concert with Dr. Elliot Fishman, Director of Johns Hopkins CT Radiology, Drs. Brian Kral and Lewis Becker spearhead this effort on behalf of the GeneSTAR team.
New work involves molecular imaging of plaque using PET under the leadership of Dr. Kral and team in Nuclear Medicine using a novel agent to identify "hot spots" of inflammation in relation to findings of plaque on CT angiography.
The ENIGMA study is combined with extensive phenotyping in nuclear perfusion imaging and exercise testing, offering a great opportunity to look at the relationship of lesions on CT angiography and functional changes. Drs. Jay Vaidya and Diane Becker are examining metabolic and traditional and nontraditional risk factors that may be mechanistically involved on various expressions of occult CAD.
Principal Investigator
Brian G. Kral, MD, MPH
Assistant Professor, Cardiology
Collaborators
Elliot K Fishman, MD
Director of Diagnostic Imaging and Body CT
Professor of Radiology and Radiological Science
Stefan Loy Zimmerman, MD
Associate Professor of Radiology and Radiological Science
Arterial Tone and Reactivity: Studies of Vascular Aging
GeneSTAR has an interest in vascular properties as a general underpinning to all of the various cardiovascular phenotypes that make up the substrate for coronary, cerebrovascular, and peripheral arterial disease.
The ARTERY study is based on the evidence that in older animals and humans, dysfunctional arterial changes including vascular stiffening and endothelial impairment are present in all vascular beds. These changes may be present at younger ages in highly susceptible individuals, predisposing them to earlier-onset cardiovascular morbidity and mortality. While arterial dysfunction increases with age, little is known about why some individuals manifest vascular impairment while relatively young and others retain preserved vascular function at older ages.
ARTERY is based on the premise that genetic variants are a significant determinant of the presence of vascular dysfunction in younger individuals and preservation of vascular function at older ages. Thus, persons with premature vascular dysfunction may have an identifiable genetic profile, more likely so in a study of a high risk population with known familial susceptibility to early-onset vascular disease.
We are examining the hypothesis that genes contribute to vascular dysfunction across a broad range of ages in GeneSTAR, a 2-generational cohort where genotyping is available. The overall goal is to study 1500 family members, including siblings of the original probands, adult offspring of the siblings or probands, and the co-parents of the offspring, for characterization of primary outcomes representing arterial functional changes, including aortic pulse wave velocity, carotid artery stiffness, and post-ischemic brachial artery flow-mediated dilatation (FMD) in the context of covariables (known atherosclerotic risk factors and inflammatory biomarkers). We are examining known variants in candidate genes and signals from the GWAS and whole genome sequencing that are associated with vascular dysfunction and markers of vascular aging. We will determine whether novel genetic loci are associated with younger age vascular dysfunction and also with successful preservation of arteries in older age individuals in families with a history of premature CAD. Such families are more likely than individuals from general population studies to be enriched with premature vascular dysfunction and also with susceptibility genes. However, family members are still sufficiently heterogeneous with regard to the vascular phenotype that genetic models can be tested. The overall objectives of this program of vascular research are to:
- determine whether there is an association between specific haplotypes or genotypes of selected mechanism-derived candidate genes and age-specific percentile levels of vascular properties, characterized by carotid artery stiffness and FMD (based on percentile for current age group), signifying the range from younger age dysfunction to successful older age vascular preservation.
- determine associations between specific haplotypes or genotypes of selected mechanism-derived candidate genes and age-specific levels of the vascular properties taking into account putatively influential covariables including the inflammatory markers (hs-CRP, IL-6, and MCP-1) and traditional atherogenic risk factors (lipids, blood pressure, glucose, obesity [body mass index]).
- determine associations of loci in the genome-wide SNP scan (excluding those SNPs in the selected candidate genes above) and age-specific percentile levels of the vascular properties, characterized by carotid artery stiffness and FMD, signifying the range from younger age dysfunction to successful older age vascular preservation, respectively.
- replicate a priority list of detected associations in two population based studies, including the Framingham Heart Study (FHS) for white subjects and the Multi-Ethnic Study of Atherosclerosis (MESA) for both white and African American subjects.
Principal Investigator
Dhananjay (Jay) Vaidya, MBBS, PhD, MPH
Associate Professor, Medicine, Epidemiology
Small Vessel Cerebrovascular Disease Cognition and Vascular Aging
Our work in brain vascular disease is represented by a series of studies in GeneSTAR participants using brain magnetic resonance imaging (3 Tesla) to examine pre-clinical small vessel cerebrovascular disease in high risk European and American families. Done in concert with the Departments of Neurology and Neuroradiology, the GeneSTAR studies are ongoing and more recently include gadolinium imaging for evaluation of the blood brain barrier and a search for novel biomarkers of premature aging in the brain, or precursors to cognitive decline. We have a particular interest in serial measures of white matter hyperintensities, brain atrophy, and functional cognitive measures.
Small vessel cerebrovascular disease (SVCD) is generally silent and is identified by white matter hyperintensities and small focal brain infarcts on magnetic resonance imaging (MRI). These lesions double the risk of subsequent stroke. We found a markedly increased prevalence of silent SVCD (73-90%) on MRI in apparently healthy middle-aged siblings of probands with known premature coronary artery disease (CAD). Siblings also underwent exercise perfusion tomography to detect occult CAD. Of subjects with occult CAD, 88% had any SVCD, and 62% had significant SVCD on MRI, while only 5.5% without occult CAD had these findings.There is a relationship between vascular disease in the brain and in the heart.
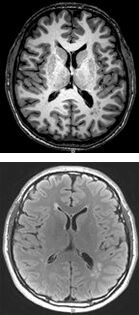
Our discovery that occult CAD and SVCD appear to co-occur in premature CAD families provided an opportunity to study models for pre-clinical small vessel disease in the brain and heart. In both clinically manifest strokes and CAD, classical risk factor models markedly underestimate incident events, suggesting that other factors, including genetic susceptibility, may be contributory. In addition, there is strong clustering of both cerebrovascular disease and CAD in families, consistent with a genetic contribution to the disease process.
To determine potential shared and unique mechanism-derived biomarkers and known risk factors related to pre-clinical vascular disease in the brain and the heart, and the contribution of attendant mechanism-specific genes, we are conducting a series of cross-sectional and longitudinal epidemiologic studies of SVCD using MRI determination of white matter hyperintensity volumes and ratios, lacunar infarcts, and occult CAD using stress sestamibi tomography of the heart as well as CT angiography.
We have used novel biological models of vascular disease to select measurements of mechanism-derived inflammatory (IL-6, TNF-α, hs-CRP, sVCAM-1, sICAM1, and MCP-1) and prothrombotic factors (PAI-1, Lp(a) and fibrinogen), endothelium dependent vascular reactivity of the brachial artery, and traditional vascular disease risk factors. More recently our work has included studies of monocytes and inflammation.
We are examining biomarkers in 800 apparently healthy 35-80 year old siblings and adult offspring from GeneSTAR families with documented premature CAD. We will also examine associations between genetic variants and SVCD using candidate genes, WGS, and GWAS.
This study is the first to examine small vessel disease in the brain and concomitantly the heart in high risk family members who are notably susceptible to coronary heart disease and cerebrovascular disease. The study also includes extensive characterization of cognitive function and measures of mental well-being, as well as comorbid events that may predispose to the neurocognitive decline associated with aging. The study is spearheaded by Dr. Paul A. Nyquist. MRI analysis support is provided by Dr. Jerry Prince, William B. Kouwenhoven Professor of Electrical and Computer Engineering, Whiting School of Engineering, Johns Hopkins University.
Principal Investigator
Paul A. Nyquist, MD
Professor, Neurology
Collaborators
Jerry L. Prince, PhD
William B. Kouwenhoven Professor Electrical and Computer Engineering
Joint appointments, Radiology and Biomedical Engineering
Norman Haughey, PhD
Professor and Vice Chair of Research, Department of Neurology
Peter C.M. Van Zijl, PhD
Department of Radiology and Radiological Science
Genetic Studies of Aspirin Responsiveness
A major GeneSTAR Research Program focus is on the role of platelets in atherothrombosis, and involves an array of genetic studies and pharmacogenomic studies. Spearheaded by Drs. Lewis Becker, Nauder Faraday, and Rehan Qayyum, this work continues to be the centerpiece of the GeneSTAR risk mechanism work. The known and hypothesized biological cascades organized and integrated into a conceptual model by our team have driven the genetic aspects of this work. (Figure1)
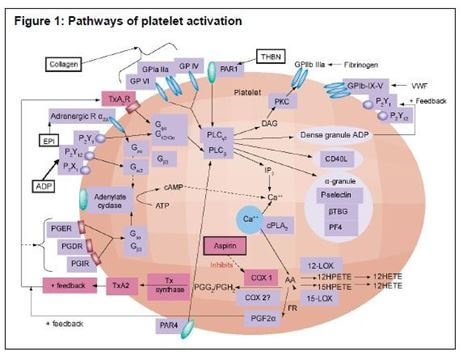
Faraday N, Becker DM, Becker LC. Pharmacogenomics of platelet responsiveness to aspirin. Pharmacogenomics 2007 Oct;8(10):1413-25. Review. PMID: 17979514
Program in Gene Environment Interactions: PROGENI
Low dose aspirin (ASA) is cost effective and efficacious for the prevention and treatment of coronary heart disease (CHD). The benefit of ASA is thought to be related to the irreversible acetylation of platelet cyclo-oxygenase-1 (COX-1), resulting in a reduction in platelet aggregation and platelet-mediated inflammation. Considerable inter-individual variation exists in the effect of ASA on platelet function, and this variability may be related to genetic variations across individuals. Our first set of genetic studies characterized the inhibitory effect of ASA on agonist-induced platelet aggregation, thromboxane and ATP release, and platelet-leukocyte conjugate formation in 3200 subjects from 650 families, European and African Americans.
Platelet function and a panel of plasma inflammatory markers (C-reactive protein, interleukin-1b, interleukin-6, monocyte chemotactic protein-1, and matrix metalloproteinase-9) were measured at baseline and after 14 days of ASA, 81 mg/day. From a list of candidate genes involved in the known biochemical pathways of platelet aggregation and platelet-mediated inflammation, genes were initially selected for genotyping of 15-20 single nucleotide polymorphisms (SNPs) per gene, based on biological importance and the presence of sufficient known SNPs in coding and/or promoter regions. After the first 1600 participants were phenotyped, a complementary genome wide scan of short tandem repeat (STR) markers and fine mapping of up to 5 regions of interest was done using SNP clusters.
Based on linkage analysis, the list of candidate genes were re-prioritized and additional genotyping was performed. ASA responsiveness is firmly established as heritable. Our primary initial findings revealed that the greatest information was coming from a variant in the PEAR-1 gene, as noted in the following publication.
Key Reference
Herrera-Galeano JE, Becker DM, Wilson AF, Yanek LR, Bray P, Vaidya D, Faraday N, Becker LC. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler Thromb Vasc Biol, 2008 Aug;28(8):1484-90. PMCID: PMC2739240. abstract full text.
Genome Wide Association Studies: STAMPEED
Aggregation of activated platelets on atherosclerotic plaques initiates thromboses of the arterial system, resulting in ischemic syndromes. The propensity of platelets to aggregate in vivo is characterized by a variety of in vitro assays. We and others have demonstrated that many of these platelet function assays are moderately to highly heritable in populations at increased risk for atherosclerosis, supporting the hypothesis that genetic variations underlie individual variability in the tendency for arterial thrombosis.
Inhibition of platelets by low dose aspirin (ASA) is also a heritable trait and genetic variations may be in part responsible for responsiveness to ASA. We have extensively characterized native platelet function and platelet function after low dose ASA (81 mg/day for 14 days) in 2000 individuals from 500 2-generational families with premature coronary artery disease (60% European American, 40% African American) participating in the Genetic Study of Aspirin Responsiveness (GeneSTAR) and the Johns Hopkins Sibling and Family Heart Study. All study participants have had a 500 short tandem repeat (STR) genome scan.
The overall goal has been to identify genes that modify the function of platelets, both under “normal native” conditions, and following low dose ASA. We performed high density single nucleotide polymorphism (SNP) genotyping (1 million SNPs, using the Illumina HumanHap550 BeadChip) covering the entire genome at an average 6kb density, on the DNA samples from the GeneSTAR participants phenotyped for platelet function.
We determined genomic loci associated with quantitative platelet phenotypes prioritized for high heritability, biological interest, and linkage to STR markers, using family based association analysis, with joint modeling of linkage and association. Phenotypes of highest priority included aggregation induced by collagen, adenosine diphosphate (ADP), arachidonic acid (AA), and epinephrine in platelet rich plasma (PRP), ATP release induced by AA, and urinary levels of the prostaglandin metabolite, 11-dehydro-thromboxane B2.
We have published our findings with the Framingham Heart Study in Nature Genetics (A. Johnson, et al), and other aspects of the work have been published and are in progress with the HAPI Heart Study of the Old Order Amish at the University of Maryland (A. Shuldiner). Work on refining the genetic findings is continuing.
Trans-Omics for Precision Medicine (TOPMed) Program
We are currently performing whole genome sequencing and collaborating with the Framingham Heart Study and the Old Order Amish Study in these efforts to identify genes related to aspirin responsiveness as part of the NHLBI TOPMed Program. Dr. Rasika Mathias is taking the TOPMed lead for GeneSTAR.
Key Reference
Johnson AD, Yanek LR, Chen M-H, Faraday N, Larson MG, Tofler G, Lin SJ, Kraja AT, Province MA, Yang Q, Becker DM, O’Donnell CJ, Becker LC. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet, 2010 Jul;42(7):608-13. PMCID: PMC3057573
Principal Investigator
Lewis C. Becker, MD
Robert L. Levy Professor, Cardiology
PARes Study
Pharmacogenomics of Anti Platelet Responses
The PARes Study has two parts. One is based on using clopidogel plus aspirin in African American families only (PARes I), and the other is based on aspirin alone in both European Americans and African Americans (PARes II).
Our goal is to find the best antiplatelet regimen for very high risk persons from a primary prevention population, those with a family history of premature coronary disease and evidence of occult disease in two vascular beds, usually the coronary arteries and the brain. Given that the responses are highly heritable, we are seeking genes that regular responsiveness to assure targeted effective therapy to assure a pro-heath risk benefit ratio. Because so little is known about coronary disease prevention in African Americans, most of the study is focused on African American families. The PARes 2 study (aspirin only) involves African Americans and European Americans.
Why Do the Study

Currently there is considerable controversy about using antiplatelet agents in primary prevention, and this study represents an effort to identify those very high-risk patients with silent preclinical disease for whom antiplatelet agents would be effective and to refine possible indications based on atherogenic pathology and platelet physiology. Through a better understanding of pharmacogenomics, we may identify subsets of at-risk primary prevention populations where demonstrable benefit favors use of antiplatelet chemoprophylaxis. Very little in particular is known about how to prevent heart attack in high risk African American families.
Biological and Genetic Background
Platelet aggregation on disrupted atherosclerotic plaques initiates vascular thrombosis, resulting in acute myocardial infarction (MI) and stroke. Aspirin (ASA) is the mainstay of primary prevention and reduces future MI risk by 32% in men, and future stroke risk by 17% in women. Failure to protect everyone at risk has been attributed in part to suboptimal platelet suppression.
ASA only inhibits a single pathway; therefore, it remains possible that in the highest risk subset of a primary prevention population, inhibition of an additional pathway will be of benefit. Clopidogrel selectively and irreversibly blocks the platelet PYreceptor pathway and might be of benefit if added to ASA in a targeted population selected on the basis of risk phenotypes and genotypes.
Recent candidate gene work in families in the GeneSTAR study, yielded a potent signal in a gene for increased native platelet aggregation as well as reduced inhibition by ASA in response to agonists for multiple pathways in European Americans. Sequencing of this gene yielded a common that was replicated in African American subjects, and in the Framingham Heart Study.
Main Goals and Protocols
The main goal is to explore the impact our strongest genetic finding on responsiveness to clopidogrel (Plavix) alone or to dual anti-platelet therapy using both ASA and clopidogrel.
We will further assess the role of other genetic variants in determining the response to single or dual anti-platelet therapy. Apparently healthy high risk subjects have been (a) identified from a proband with early-onset CAD , (b) genotyped on the Illumina 1 million platform, with imputation to 2.5 million SNPs, and (c) fully phenotyped for occult vascular disease in the brain using MRI and heart using CT angiography.
We plan to characterize the variance in platelet aggregation to multiple agonists after therapy with clopidogrel alone and therapy with clopidogrel and ASA in a subjects having both significant occult CAD identified by coronary CT angiography, and small or large vessel cerebrovascular disease identified by cranial MRI/MRA. We further plan to determine the extent to which variants identified modify platelet responsiveness to inhibition by clopidogrel alone and in combination with ASA in this high-risk subset. In addition, our aim is to determine the extent to which variants in other recently discovered genes, by themselves, and in combination with our known gene finding, modify platelet responsiveness to clopidogrel alone and with ASA in this high-risk subset. Lastly, we also want to determine what changes in platelet mRNA are produced by aspirin alone and by aspirin with clopidogrel.
Pharmacogenomics and Tailored Medicine

Currently there is controversy about using antiplatelet agents in primary prevention, and this study represents an effort to identify high-risk seemingly healthy people with silent preclinical disease for whom antiplatelet agents would be effective. The study is also designed to refine possible medication indications based on platelet physiology.
Through a better understanding of pharmacogenomics, we may identify subsets of at-risk primary prevention populations, particularly among high risk African Americans, where demonstrable benefit and low risk favors the use of one or more antiplatelet agents for chemoprophylaxis, particularly in myocardial infarction and thrombotic stroke.
GeneSTAR has a longstanding history of studying the pharmacogenomics of antithrombotic therapies for primary prevention in high risk populations, and in PARes I is focusing on African American families because so little has been done to understand the benefits of these agents in this population which has a particularly pro-aggregable platelet profile, and high risk for acute thrombotic events. Funded in part by the NHLBI.
The study involves Dr. Rehan Qayyum as the Principal Investigator, Drs. Lewis Becker, Diane Becker, Jay Vaidya, Rasika Mathias, Nauder Faraday, and Brian Kral
Principal Investigator
Rehan Qayyum, MD, MPH
Assistant Professor, Medicine (Adjunct)
Professor, Medicine
Virginia Commonwealth University
