Researchers examine every facet of the digestive system to find better ways to treat illnesses and conditions that originate in the gastrointestinal system, liver and pancreas, from genetic biomarkers and molecular pathogenesis to the role of alcohol consumption and nutrition.
In This Section
-
GI & Hepatology Clinical and Translational Research Unit
JHED required

Research in Action
At the Johns Hopkins IBD Center, groundbreaking work is being done. For the past three years, Dr. Alyssa Parian and her colleagues - as part of a three year grant - have been developing solutions for perianal fistulas.
Featured Research and Patient Care Stories
-
Johns Hopkins Study Reveals Racial and Socioeconomic Disparities in Liver Transplant
Researchers demonstrate that patients from underrepresented populations living in low-socioeconomic neighborhoods are at significant risk of not receiving the care they require.

-
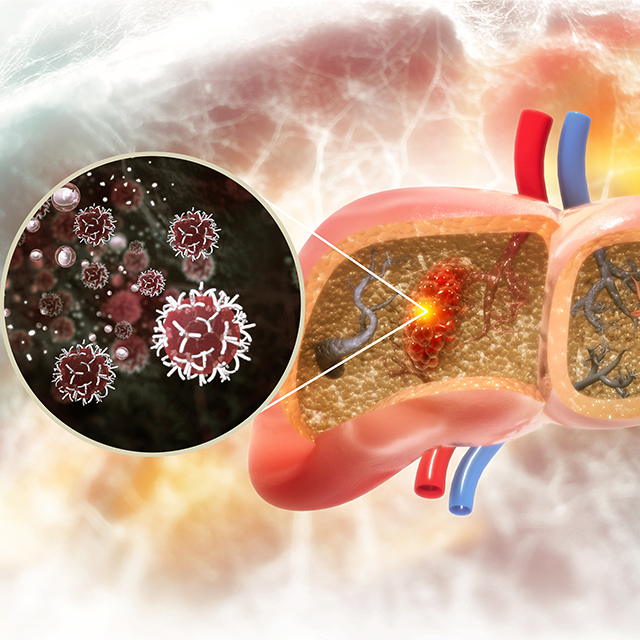
Johns Hopkins Hepatologists Detect Liver Cancer in Urine Test
Researchers show that screening urine for ctDNA as part of a two-stage test increases detection sensitivity from 40% to 77% in early-stage liver cancer, and from 62% to 92% in the very early stage of disease.
-
Johns Hopkins Among Few in U.S. to Offer Early Liver Transplants for Patients with Alcohol-Associated Liver Disease
New research opens window into understanding the needs of patients after the procedure.
Research Funding
Initially established with the generous contribution of Harvey and Lyn Meyerhoff, the majority of current research funding in basic science comes from investigator-initiated independent grant support from the NIH. Funding is also provided by major national organizations such as the American Gastroenterology Association, American Liver Foundation, American Heart Association, American Cancer Society, Crohn’s and Colitis Foundation of America and a variety of private sources including major pharmaceutical firms.




